All News & Updates

MLDeCNV: A machine learning approach for predicting copy number variation types in plant genomes
Highlights •MLDeCNV is a novel open-source CNV prediction tool optimized for plant genomes. •Built on the XGBoost model, using 32 features from coverage metrics, nucleotide composition, and sequencing statistics. •Robust with 89.27 % accuracy, 89.3 % precision, recall, and F1-score and 0.9783 AUC. •Outperforms traditional ML models and CNV detection tools. •Addresses the growing need for identifying hidden heritability linked to plant CNVs. https://www.sciencedirect.com/science/article/pii/S0010482525017482
CRISPR-GATE: A One-Stop Repository and Guide to Computational Resources for Genome Editing Experimentation
https://apps.crossref.org/pendingpub/pendingpub.html?doi=10.1093%2Fbib%2Fbbaf575
Genome-wide association study reveals putative candidate genes in cucumber (Cucumis sativus L.) associated with shoot and root system architecture traits under water stress conditions. BMC Plant Biology
https://bmcplantbiol.biomedcentral.com/articles/10.1186/s12870-025-06875-2
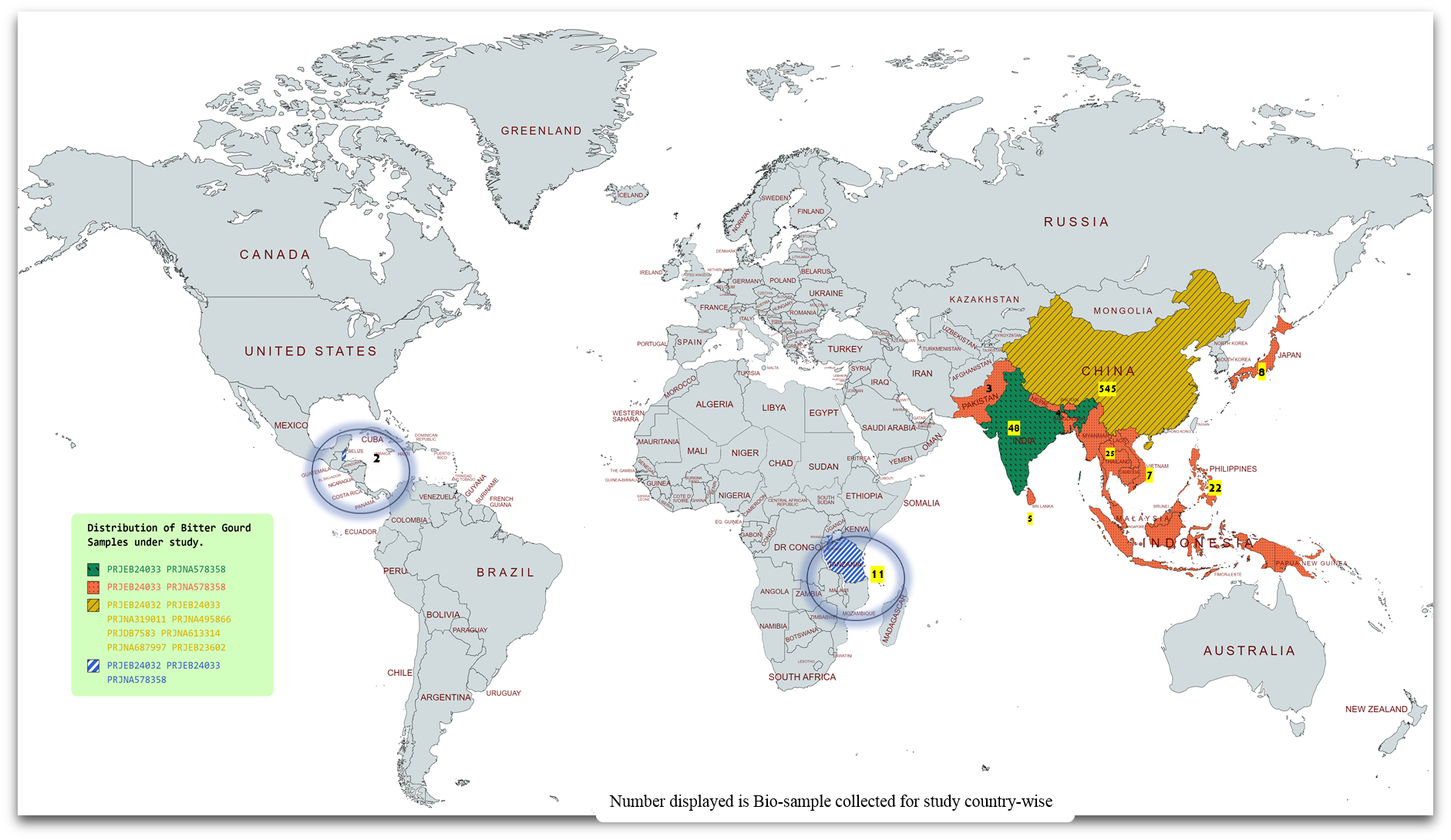
BgDB: a comprehensive genomic resource information system of bitter gourd for accelerated breeding programme
Bitter gourd, scientifically known as Momordica charantia L. with 2n = 22, is a widely recognized medicinal vegetable, renowned for its multifaceted health benefits, primarily acclaimed for its lipid- and glucose-lowering effects. Its growing demands as a food source and for industrial applications necessitate value addition in ongoing breeding initiatives to enhance genotypic traits in multifarious ways. A thorough understanding of the underlying molecular footprint is warranted for characterization, which still remains underexplored relative to other cash crops. Though a chromosome-level genome assembly of bitter gourd is available, scattered and fragmented information becomes an obstacle for assisted breeding and gene editing. Therefore, it is crucial to further dissect structural and molecular variants, noncoding RNAs (ncRNAs), transcription factors, and transcripts from whole-genome and resequencing projects. The present study leads to the development of a comprehensive genomic resource, BgDB (Bitter Gourd Resource Database) at a single platform, vital for advanced bitter gourd breeding programmes for raising bitter gourd varieties with traits of significant social and economic value. BgDB, available at https://bgdb.daasbioinfromaticsteam.in/index.php, is a user-friendly, three-tier database that offers a comprehensive interface with detailed analysed information, including 114 598 transcripts, 4914 differentially expressed genes, 32 570 predicted simple sequence repeat markers, and 162 850 primers for downstream applications. It also catalogues extensive annotations of bitter gourd-specific single nucleotide polymorphisms/insertions and deletions, long noncoding RNAs, circular RNAs, microRNAs, 1220 transcription factors, 295 transcription regulators, and 146 quantitative trait loci (QTL) distributed throughout the chromosomes. This genomic resource is poised to significantly advance genetic diversity analyses, population and varietal differentiation, and trait optimization. It further facilitates the exploration of regulatory ncRNA elements, key transcripts, and essential transcription factors and regulators. The discovery of QTL will aid in the development of improved bitter gourd varieties in the endeavour of enhanced productivity. Beyond comprehensive datasets, the future integration of multi-omics resources could profoundly advance and fully unlock the potential of databases. Database URL: https://bgdb.daasbioinfromaticsteam.in/index.php.
Carabeef: Unexploited Healthy Meat from Climate-Resilient Water Buffaloes for Global Nutritional Security
Buffaloes (Bubalus bubalis) are versatile and adaptable ruminants on account for providing meat, milk, draught power, transportation, on-farm labour, and manure in several developing nations. Buffalo’s adaptability to the fast conversion of poor-quality pasture into biomass makes it a lucrative meat choice. Buffalo’s meat (carabeef) is appraised as a pivotal food source as it is profoundly packed with superior nutritional, technological, and organoleptic attributes in comparison to other ruminant’s meat. Strikingly, it offers a myriad of bioactive compounds with multifaceted actionable wellness resources, successively opening a roadmap for interventions on personalized nutrition. Various conventional and pragmatic approaches, such as marination, enzymatic tenderization, and many others, have been employed to improve meat quality. These methods primarily focus on the programmable manipulation of the buffalo diet pattern, the impact of cooking and processing to enhance shelf-life, fostering an effective value chain for the development of value-added products, and providing exceptional consideration as an alternative diet for captive animals. The buffalo meat trade is opportunistic and prosperous in nature, with several linked bottlenecks. Frozen meat paves the way by bridging the gap for the buffalo meat trade across the world. It is further processed into value-added meat products and by-products, which are an attractive choice for consumers and trade. We have formulated efficient technical guidance to maintain frozen meat, and the way forward to value addition has been highlighted. Effective utilization of abattoir meat by-product is promising and sustainable in terms of both accrual and environmental considerations. There are various health benefits, including satiety, which helps to control appetite, and weight management through the consumption of balanced proportions; however, foresight must be exercised before consuming unprocessed meat. The development of recent genomics and biotechnological trends comes into light to overcome adulteration practices often prevalent in the meat industry, opening new avenues to effloresce economic potential of buffalo meat. https://link.springer.com/article/10.1007/s11947-025-03906-6#Abs1
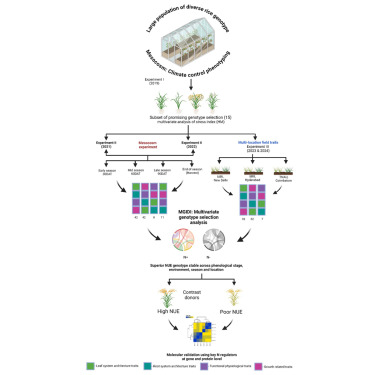
Identification of superior rice donors with enhanced nitrogen use efficiency using a comprehensive multivariate genotype selection strategy
Improving nitrogen use efficiency (NUE) of rice plants utilizing a few end-of-season traits poses a severe phenotyping bottleneck in exploring the genetic diversity of a large population and genotype selection accuracy. Therefore, a comprehensive multivariate genotype selection strategy was developed to explore maximum genetic variation of 300 diverse rice genotypes and accurately select promising rice donors with enhanced NUE traits on a multi-year (2019, 2021, and 2022) -trait (126 traits) -environment (2) -temporal (5) –location (3) scale. The multi-trait genotype ideotype distance index (MGIDI) ranked Cauvery, Suweon, RPW9-4 (SSI) and BAM3690 (IC463705) as superior NUE genotypes; Moroberekan, PUSA1121 and BAM8315 (Basmati 370) as low NUE genotypes. The multi-location field performance and molecular analysis of key nitrogen assimilatory genes confirmed the outperformance of the Cauvery genotype in terms of possessing efficient N sensing, uptake, transport and assimilation characteristics under N-limited conditions. Phenome-wide multivariate analysis highlights root-shoot plastic response as a key target trait for breeding rice genotypes resilient to N stress conditions. https://www.cell.com/iscience/fulltext/S2589-0042(25)01541-X
Interrelationships of morpho-physiological, biochemical traits, and multivariate analysis in a diverse pepper (Capsicum annuum L.) panel under high temperature conditions using a multi-trait mixed model
In the present study 149 diverse Capsicum annuum L. genotypes were evaluated for 22 different morpho-physiological and biochemical traits for high temperature tolerance across two consecutive summer seasons under open-field conditions. Both individual and pooled season analysis of variance (ANOVA) revealed significant genotypic variances for all traits, indicating substantial genetic variation within the diversity panel. Genotype × Environment interactions were significant for traits such as number of primary branches (NPB), leaf length (LL), average fruit width (AFW), average fruit weight (AF Wt), hundred seed weight (HSW), canopy temperature (CT), canopy temperature depression (CTD), pollen viability (PV), net photosynthetic rate (Pn), and catalase activity (CAT), suggesting differential responses of genotypes to environmental conditions. Descriptive statistics and ANOVA indicated an enormous amount of variability for all traits. Wilcoxon statistic comparisons revealed significant mean differences between seasons for all traits except leaf width (LW), PV, Pn, malondialdehyde level (MDA), and CAT. Additionally, most of the studied morpho-physiological and biochemical traits presented high heritability and genetic advance as a percentage of the mean, genotypic coefficient of variation (GCV), and phenotypic coefficient of variation (PCV), suggesting that additive gene impact predominates; henceforth, selection would be effective for their improvement. We also found significant positive correlations between fruit yield per plant (FYP) with PV, number of fruits per plant (NFPP), AF Wt, CTD, average fruit length (AFL), AFW, number of seeds per fruit (NSPF), Pn, stomatal conductance (Gs), transpiration rate (Tr), intercellular CO2 concentration (Ci), CCI, and antioxidant enzymes (SOD [superoxide dismutase activity] and CAT). Besides, principal component (PC) analysis revealed that the first seven PCs explained 81.62% of the total variation, and the Multi-trait Genotype-Ideotype Distance Index (MGIDI) selected 22 promising pepper genotypes for heat tolerance breeding. Notably, eight of them had high PC scores. Therefore, these identified genotypes can serve as most valuable genetic resources to develop heat-tolerant pepper cultivars. https://link.springer.com/article/10.1007/s10722-025-02551-4

IARI Tops NIRF Ranking in Agriculture and Secures 2nd Rank in SDG Institute Category
The ICAR-Indian Agricultural Research Institute (IARI), popularly known as Pusa Institute, has once again brought laurels to the nation by excelling in the National Institutional Ranking Framework (NIRF) 2025 announced by the Ministry of Education, Government of India. Shri Dharmendra Pradhan, Hon'ble Education Minister, Government of India presented the NIRF ranking certificates on 4th September, 2025. IARI has secured the 1st position in the Agriculture and Allied Sectors category, reaffirming its premier status in agricultural education, research, and innovation. In the newly introduced Sustainable Development Goals (SDG) category, the institute made an impressive debut by achieving the 2nd rank, just behind IIT Madras, showcasing its strong contributions towards sustainability, inclusivity, and national priorities. In the Overall category, IARI was ranked 24th, while in the Research Institutions category it stood at 29th, marking a remarkable improvement of eight positions compared to last year. This achievement underscores IARI’s growing national relevance and global recognition across stringent evaluation parameters such as teaching, learning and resources, research and professional practice, graduation outcomes, outreach, and inclusivity. At the award ceremony, Dr. Ch. Srinivasa Rao, Director and Vice Chancellor of IARI, and the Dean of the Institute Dr Anupama Singh, received the recognition on behalf of IARI. On this occasion, both of them congratulated the scientists, staff, and students of the institute for their hard work, dedication, and collective efforts that have propelled IARI to newer heights. Expressing his happiness, Dr. Ch. Srinivasa Rao said: “This recognition is a testimony to the scientific excellence, innovation, and teamwork of IARI. It reflects the institute’s vision of contributing to national food and nutritional security, sustainable agriculture, and the global leadership of India in agricultural sciences.” He further added: “Our consistent rise in NIRF rankings is the result of relentless dedication by faculty, staff, and students. It inspires us to further strengthen our teaching, research, and extension systems to meet the aspirations of farmers and the nation.” This recognition in the NIRF 2025 Rankings not only strengthens IARI’s position as the country’s foremost agricultural institute but also underscores its commitment to shaping the future of farming, empowering farmers, and achieving the goals of Viksit Bharat.
